ELISA Guide; Part 1: Introduction to ELISA, Formats and Signal Amplification

Introduction to ELISA
Enzyme-linked immunosorbent assay (ELISA) was first described in 1971 when it was developed as a safer and more convenient alternative to radioimmunoassay. It has since become a widely used technique for detecting and, most importantly, quantifying one or more specific proteins within a complex mixture.
The main advantages of ELISA are that it is quick and easy to perform and can be adapted to detect almost any analyte. ELISA is also incredibly sensitive, affords ample opportunities for multiplexing, and has the capacity to handle large sample numbers.
Although several ELISA formats have evolved since the earliest experiments were performed, the most popular include direct detection of a pre-coated analyte on a plate by a primary antibody in conjunction with a labeled secondary. Another popular format is a sandwich assay, where the analyte is captured at the plate surface by a pre-coated primary antibody which is subsequently detected by another primary and labeled secondary pair. Irrespective of the chosen format, the success of any ELISA depends on high-quality antibodies and accessory reagents, and meticulous attention to detail during assay development and validation.
The aim of this guide is to provide a general overview of the ELISA technique, highlighting the many ways that Jackson ImmunoResearch can support your ELISA-based studies. Also included is information about assay optimization, data analysis, and troubleshooting, to help you generate results you can trust. Whether you’re running an ELISA for the very first time, or you’re looking for ways to enhance an existing ELISA workflow, Jackson ImmunoResearch is here to help.
Covered in this section
ELISA Formats
Over the past 50 years, several different ELISA formats have been developed. These are categorized as direct, indirect, sandwich and competitive ELISA, with the chosen method largely being dictated by the aims of the study.
Direct ELISA
In a direct ELISA, the analyte is bound directly to the microplate wells and is then detected with a reporter-conjugated antibody specific to the target. This is shown in Figure 1.
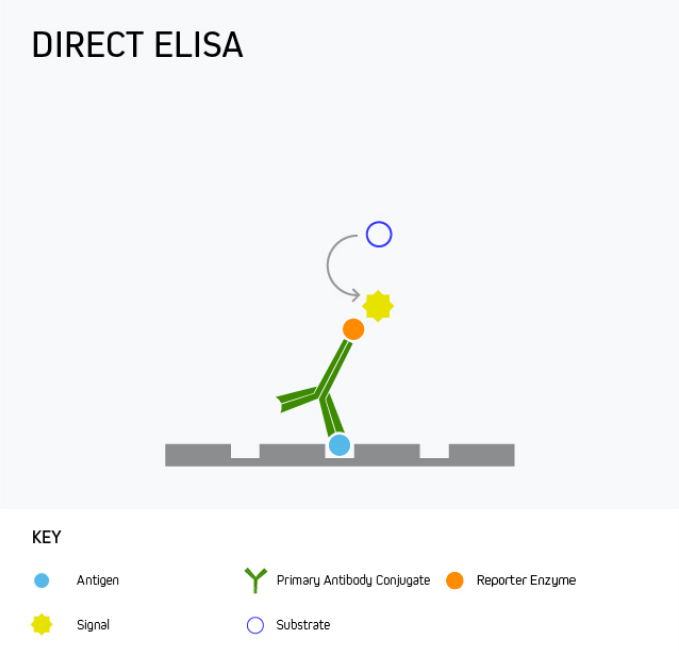
A direct ELISA offers the advantage of being quick and easy to run since it involves little more than coating the microplate with the analyte, blocking, and then adding the detection antibody. However, it has only limited sensitivity as there is no amplification from a secondary antibody, meaning alternative formats are often preferred.
One use for direct ELISA is to detect a specific protein in patient serum. This is achieved by coating the microplate wells with the serum sample, then adding the labeled detection antibody.
Indirect ELISA
During an indirect ELISA, the analyte is bound directly to the microplate wells before being detected using an analyte-specific antibody in complex with a reporter molecule-conjugated secondary antibody. This is shown in Figure 2.
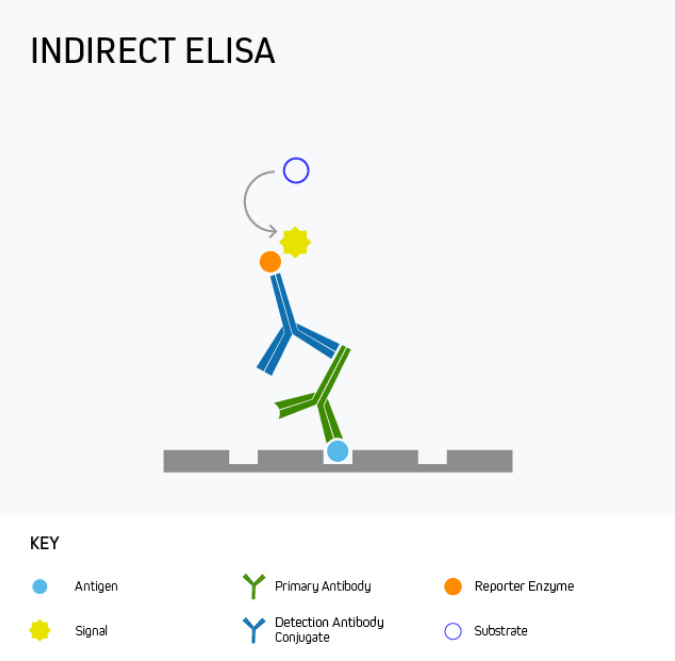
The main advantage of an indirect ELISA is that it provides greater sensitivity than a direct ELISA because multiple secondary antibodies can bind the analyte-specific antibody. It is also more flexible since directly conjugated antibodies specific to the target analyte are not always available or can be expensive. Disadvantages of indirect ELISA are that it involves an extra protocol step compared to a direct ELISA and introduces the potential for cross-reactivity between secondary antibodies and the target analyte.
Indirect ELISA is mainly used for measuring the antibody concentration in a sample, for example, to quantify antibody production in response to vaccination with a recombinant form of the SARS-CoV-2 SPIKE protein, or during allergy testing to detect the presence of antibodies in serum following treatment with a potential allergen such as peanut oil.
Sandwich ELISA
Sandwich ELISA is the most commonly-used ELISA format, with almost unlimited utility for basic research as well as for clinical applications like disease diagnosis and health monitoring. This format uses matched antibody pairs for capture and detection. First, an analyte-specific capture antibody is bound directly to microplate wells which are subsequently blocked. The analyte-containing sample is then added, followed by a second analyte-specific primary antibody which may, or may not be labeled with a reporter enzyme. In cases where the second primary antibody is not labeled, a labeled secondary must be employed to generate an output signal.
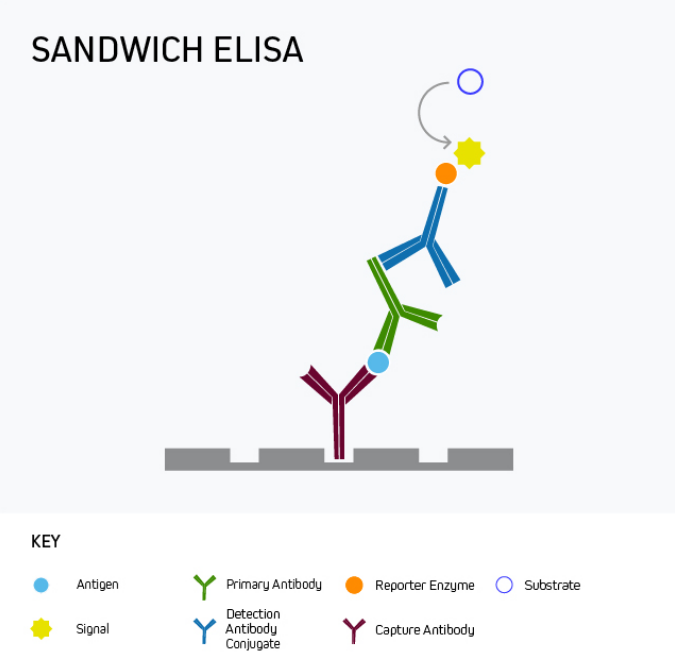
Sandwich ELISA benefits from superior specificity compared to either a direct or indirect ELISA because two distinct analyte-binding antibodies (each recognizing a different antigenic epitope) are used. It may also provide greater sensitivity, and even low-abundance antigens can be detected within complex samples such as serum. These advantages of sandwich ELISA far outweigh its main drawback, namely that it involves more protocol steps than direct or indirect ELISA.
It is important to remember that a sandwich ELISA relies on using a matched antibody pair that has been rigorously validated for the ELISA application. Not only is this critical to prevent the two antibodies competing with one another to bind the target epitope, but it also avoids the potential for inter-antibody cross-reactivity that can generate unwanted background signal by introducing non-specific binding.
Competitive ELISA
Any of the ELISA formats just described can be converted to a competitive ELISA. In a competitive ELISA, the target analyte (either a protein or an antibody) competes with a reference for binding to a known amount of labeled antibody or protein. The higher the concentration of the target analyte, the lower the output signal. This is shown in Figure 4.
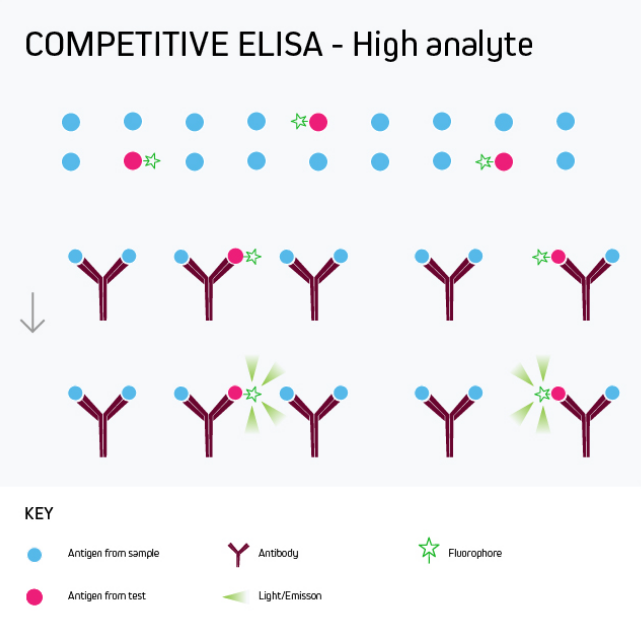
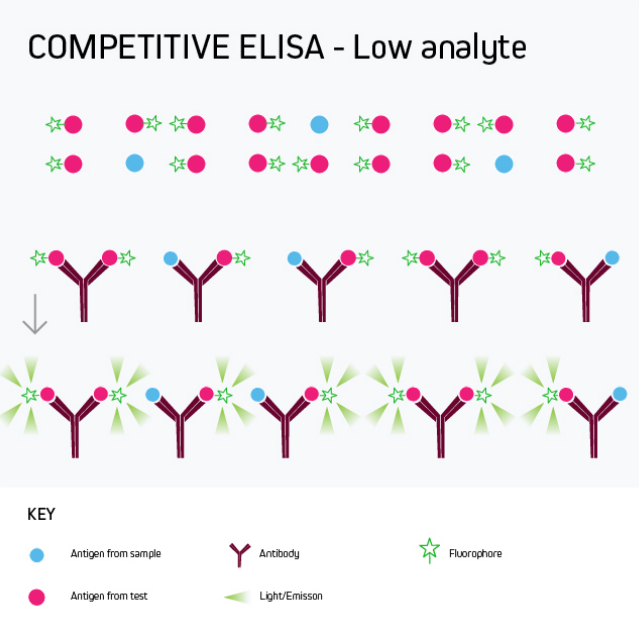
A competitive ELISA determines the concentration of an antigen or an antibody in a sample by measuring signal interference. Essentially, the target of interest competes with a reference for binding to a known amount of a labeled binding partner, with a high target concentration yielding a low signal. This type of ELISA is the most challenging to set up, especially where the competing biomolecule must be labeled in-house.
Competitive ELISA is typically used when only one antibody is available for the target analyte or when the target analyte is small and cannot be bound by two different antibodies simultaneously. Its main applications include testing blood taken from professional athletes for the presence of banned drugs and monitoring environmental samples such as river water for chemical contamination.
Signal amplification
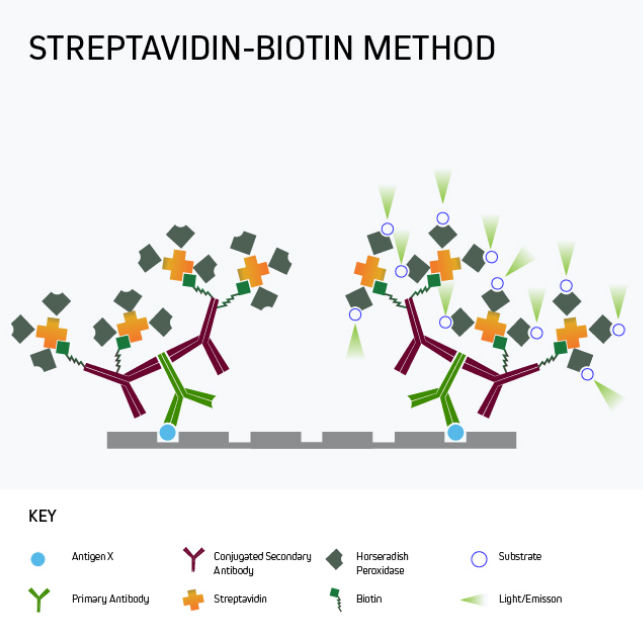
In many situations, especially where the target analyte is present in samples at only low levels, it may be necessary to amplify the ELISA signal. A popular approach is to use labeled streptavidin-biotin-complex (LSAB) signal amplification, a technique that exploits the high-affinity streptavidin-biotin interaction to add multiple detection moieties to the target analyte.
Figure 5 shows a form of signal amplification using biotinylated secondary antibodies to bind the analyte-specific antibody after the primary antibody has reacted with the plate-bound analyte. By detecting this interaction with labeled streptavidin, signal amplification is achieved by virtue of 1) multiple biotins being attached to each secondary antibody and 2) the tetrameric nature of streptavidin, which has four biotin-binding sites per molecule. It can clearly be seen that the number of detection moieties increases in the final complex.
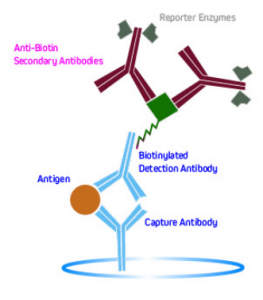
Another method is to use labeled anti-biotin antibodies to detect biotinylated primaries. This is shown in Figure 6.
| Learn more: | Do more: |
|---|---|
| Colorimetric western blotting | Spectra Viewer |
| Chemiluminescence western blotting | Antibodies for signal enhancement |
| Fluorescent western blotting | |



