
Surface Plasmon Resonance (SPR) is a label-free optical biosensing technique that allows researchers to measure molecular interactions in real-time. It has been widely adopted for drug discovery, where it is used to investigate events such as the binding of small molecules to therapeutic protein targets and the process of antibody-antigen recognition.

SPR principles
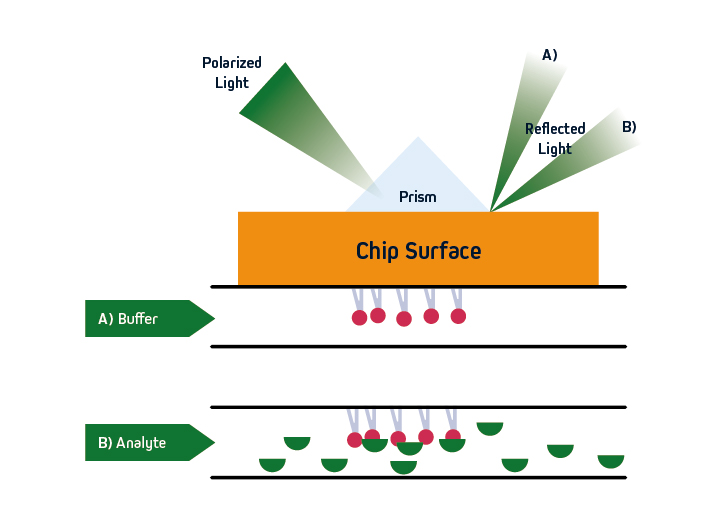
SPR is based on the concept of total internal reflection (TIRF), whereby light that is traveling through an optically dense medium (e.g., glass) reaches an interface with a less optically dense medium (e.g., liquid) and is reflected back.
Specifically, in SPR instruments such as Cytiva’s Biacore™ systems, the incident light is polarized and the interface comprises a sensor that is coated with an electrically conducting metal film (typically gold due to its stability). When the polarized light hits the sensor surface, an energy transfer takes place, generating electron charge density waves known as plasmons. This reduces the intensity of the reflected beam, which is measured on a detector at a specific angle (the resonance angle).
By immobilizing a ligand on the sensor, and exposing it to an analyte that is free in solution, the refractive index close to the sensor surface changes upon analyte binding. The resultant shift in resonance angle is proportional to the mass on the sensor surface, allowing for monitoring of molecular interactions in real-time. Importantly, because the light does not penetrate the sample, SPR is compatible with complex sample types including serum and pathogenic material. The basic principles of SPR are shown in Figure 1.
Binding phases – the sensorgram
A typical SPR experiment begins with inserting the sensor into the instrument and priming the microfluidic system with buffer. The ligand (usually a protein) is then injected and becomes immobilized on the sensor surface, either via covalent coupling or with a capture-based method. Dextran-based sensors are most often used, although the sensor type and immobilization conditions (e.g., type of coating, ligand concentration, buffer composition, pH) should always be optimized for the type of interaction being studied. Next, analysis is initiated and the data is provided in the form of a sensorgram. This displays time on the x-axis and the binding response on the y-axis, as shown in Figure 2.
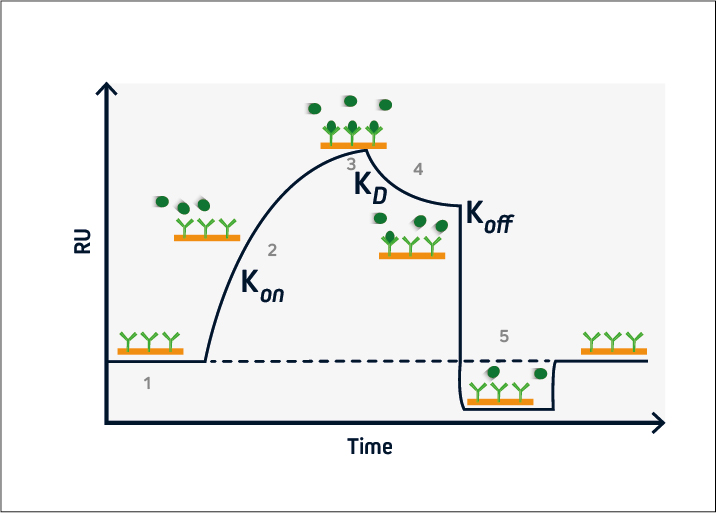
During the baseline phase (1), buffer flows across the sensor surface, conditioning the sensor and allowing any instabilities to be observed. This is critical to ensure a stable baseline and produce accurate results. The association phase (2) is initiated at t = 0, when the analyte is introduced into the system. Analyte binding at the sensor surface leads to an increase in signal, from which the association rate (kon) can be determined.
At steady-state (3), the amount of analyte associating with and dissociating from the ligand is equivalent, and the equilibrium constant (KD) can be calculated. This is followed by the dissociation phase (4), when the analyte solution is replaced with buffer and the dissociation rate (koff) can be established.
If the ligand-analyte complex has a long half-life, a regeneration phase may be necessary (5). This involves adding a solution to disrupt the ligand-analyte interaction and restore the signal to the original baseline. Solutions with a high salt concentration or low pH are frequently used for sensor regeneration.
SPR applications
In addition to being used for studying binding kinetics, SPR allows researchers to address many other facets of biomolecular binding. These include how specific an interaction is, whether a ligand has high or low affinity for its analyte, and what the biologically active concentration of a given molecule is within a sample. In addition, SPR allows for investigation of how these parameters can be manipulated.
Published applications of SPR include its use for fragment-based screening, kinetic evaluation of kinase inhibitors, and characterization of lipid-protein interactions. SPR is also cited for streamlining the development of antibody-drug conjugates (ADCs), performing quality assessment of bispecific antibodies, and measuring the kinetics of PROTAC ternary complexes1-6. The use of SPR continues to grow as researchers become more familiar with the technology.
References
- Chavanieu A, Pugnière M. Developments in SPR Fragment Screening. Expert Opin Drug Discov. 2016;11(5):489-499. doi:10.1517/17460441.2016.1160888/
- Kitagawa D, Gouda M, Kirii Y. Quick evaluation of kinase inhibitors by surface plasmon resonance using single-site specifically biotinylated kinases. J Biomol Screen. 2014;19(3):453-461. doi:10.1177/1087057113506051/
- Stahelin RV. Surface plasmon resonance: a useful technique for cell biologists to characterize biomolecular interactions. Mol Biol Cell. 2013;24(7):883-886. doi:10.1091/mbc.E12-10-0713/
- Healey GD, Frostell A, Fagge T, Gonzalez D, Conlan RS. A RAGE-Targeted Antibody-Drug Conjugate: Surface Plasmon Resonance as a Platform for Accelerating Effective ADC Design and Development. Antibodies (Basel). 2019;8(1):7. Published 2019 Jan 7. doi:10.3390/antib8010007/
- Gassner C, Lipsmeier F, Metzger P, et al. Development and validation of a novel SPR-based assay principle for bispecific molecules. J Pharm Biomed Anal. 2015;102:144-149. doi:10.1016/j.jpba.2014.09.007/
- Roy MJ, Winkler S, Hughes SJ, et al. SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem Biol. 2019;14(3):361-368. doi:10.1021/acschembio.9b00092/
| Learn more: | Do more: |
|---|---|
| Indirect and direct Western blotting | Exhibition Schedule |
| Chemiluminescence western blotting | Cite and Win |
| An Introduction to Expansion Microscopy | Past Competition Winners |



