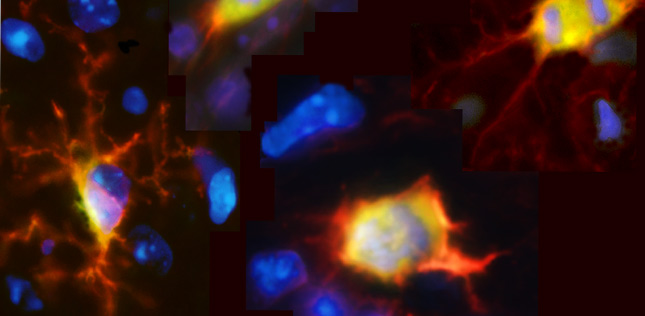
Spatial biology is a rapidly evolving field of research that puts cellular information into its native 2D or 3D framework with a broad range of experimental techniques. By allowing researchers to study proteins, RNA transcripts, and other analytes across the tissue landscape, spatial biology can provide novel insights into the biological systems governing human health and disease, which could ultimately lead to the development of better treatments for cancer, neurodegeneration, and other disorders.
What is spatial biology?
Put simply, spatial biology is the mapping of biological molecules within intact tissue samples. It encompasses methods for visualizing proteins or RNA transcripts, as well as techniques for spatially-resolved DNA sequencing, many of which can be combined for multimodal analysis. Critically, because spatial biology can reveal which cells are present in a tissue, which biomarkers they express, and how different cell types interact with one another to influence the tissue microenvironment, it puts cellular function into the context of the native tissue architecture.
What techniques are used for spatial biology?
Established techniques for spatial biology include basic immunohistochemistry (IHC), which uses antibodies labeled with enzymes or fluorescent dyes for detecting proteins of interest; single-molecule fluorescence in situ hybridization (smFISH), which relies on labeled DNA oligo probes for visualizing specific DNA sequences; and RNA in situ hybridization (RNA ISH), a method analogous to smFISH that is used for localizing RNA transcripts. However, because these techniques have historically been limited to detecting just a handful of targets, more advanced technologies have been developed to investigate spatial biology at a much deeper scale. While it would be impossible to cover all of the available approaches here, the following are some leading examples:
Spatial proteomics
Tissue-based cyclic immunofluorescence (t-CyCIF)
First reported in 2015, cyclic immunofluorescence (CycIF) is a technique that involves repeated rounds of immunofluorescence staining and fluorophore inactivation (using a high pH hydrogen peroxide solution in the presence of light) to achieve highly multiplexed imaging of proteins in single cells1. It was adapted to a tissue-based method in 2018 by the authors of the original publication, who showed t-CyCIF to be capable of tracking over 60 different targets in tissue samples from human patients2. A main advantage of both CycIF and t-CyCIF is that they use commercially available antibody reagents and conventional optical microscopes, making them widely accessible to researchers.
Imaging Mass Cytometry™ (IMC™)
Imaging Mass Cytometry, based on CyTOF® technology, uses antibodies labeled with metal tags for detecting protein targets of interest. Initially described in 2014, it involves immunostaining the sample and introducing it into a specialized imaging system, where it is ablated by a UV laser, one pixel at a time, before being transferred to an inductively coupled plasma for ionization3. The ions are then quantified and the signals corresponding to each tag are correlated back to the respective markers to generate an image. Compared with t-CyCIF, Imaging Mass Cytometry is more efficient as it enables the simultaneous imaging of 40 or more different targets. However, the need for dedicated antibody reagents and instrumentation can limit its accessibility.
Co-detection by indexing (CODEX)
CODEX, a method developed in 2018, uses DNA-barcoded antibodies for protein detection4. Like t-CyCIF, it involves iterative cycles of staining and imaging, although antibody binding events are visualized using fluorescent dNTP analogs and an in situ polymerization-based indexing procedure. In the original study, CODEX was successfully used to detect up to 66 antigens in frozen tissue, although the procedure is reported to have an unlimited multiplexing capacity and has since been adapted for use with formalin-fixed paraffin-embedded (FFPE) tissue samples. CODEX can be performed on most three-color fluorescence microscopes with a motorized stage, using antibodies labeled with uniquely designed oligonucleotide duplexes to enable iterative stepwise visualization.
Spatial transcriptomics
Sequential fluorescence in situ hybridization (seqFISH)
seqFISH was developed in 2014 as a technique for profiling mRNA transcripts in single cells5. It involves sequential rounds of probe hybridization, imaging, and stripping (using DNase I), each time using the same set of probe sequences, but coupled to different fluorophores, in order to generate a barcode. The individual barcodes can then be quantified to provide the abundance of the corresponding transcripts in each cell. An evolution of seqFISH, known as seqFISH+, overcomes challenges related to optical crowding, which occurs when too many transcripts within a densely packed cell are simultaneously labeled and imaged6. Using seqFISH+, it is possible to image mRNAs for as many as 10,000 genes in single cells with high accuracy and resolution.
Multiplexed error-robust FISH (MERFISH)
MERFISH is based on similar principles to seqFISH, but uses an error-robust barcoding scheme for detecting mRNA species7. Explicitly, each target gene is assigned a unique binary barcode, consisting of a sequence of zeros and ones, and fluorescent probes encoding the various barcodes are used for target visualization via sequential rounds of smFISH. This approach ensures that any errors accumulated through multiple imaging rounds can be detected and/or corrected, providing greater confidence in results.
RNAscope
First described in 2012, RNAscope is an ISH technology that allows for simultaneous signal amplification and background suppression through the use of novel probes8. Specifically, pairs of probes, each containing an 18- to 25-base region complementary to the target mRNA but with a different 14-base tail sequence, are hybridized to the transcript of interest, then the two tail sequences together form a 28-base hybridization site for a preamplifier, allowing for amplification. Because it is highly unlikely that a non-specific hybridization event will result in the formation of the 28-base hybridization site, and because a single 14-base tail sequence is unable to bind the preamplifier sufficiently to enable signal amplification, background is suppressed. The probes can be either fluorescently labeled for visualization with a fluorescent microscope or conjugated to alkaline phosphatase (AP) or horseradish peroxidase (HRP) for chromogenic reactions.
Spatial genomics
Unlike spatial proteomics and spatial transcriptomics, the term spatial genomics is less clearly defined. It can be used to describe how the genome sequence varies across different regions of a tissue or organ. For example, a method known as multiregion sequencing allows for studying genomic heterogeneity through exome sequencing, chromosome aberration analysis, and ploidy profiling on multiple spatially separated samples9. Alternatively, spatial genomics can refer to how the genome is organized within the cell nucleus. Methods used for this approach include high-throughput chromosome conformation capture (Hi-C), which maps the three-dimensional architecture of whole genomes by coupling proximity-based ligation with massively parallel sequencing; DamID, which tethers E. coli DNA adenine methyltransferase (Dam) to a chromatin protein for genome-wide mapping of chromatin targets; and Genome Organization using CUT and RUN Technology (GO-CaRT), which allows for mapping the genome relative to non-membranous nuclear compartments10,11,12.
Spatial multiomics
Many of the techniques just described, and others besides, can be combined for multimodal analysis. For example, RNAscope probes have been labeled with heavy metals and used for IMC in conjunction with metal-labeled antibodies to enable transcriptomic and proteomic interrogation of breast cancer tissue samples13. And, more recently, a method termed Multiplexed Imaging of Nucleome Architectures (MINA) has been developed that combines chromatin tracing, MERFISH, and multiplexed protein imaging into a single technology14. In addition, a growing number of commercial platforms is entering the market to support different combinations of ‘omics readouts.
Conclusion
Spatial biology represents a powerful new field of research that promises to resolve many unanswered biological questions. Whether you want to determine how chromosome conformation influences normal embryonic development, or you need to combine spatial proteomic data with transcriptome analysis to better understand the dynamics of the tumor microenvironment, there is a spatial biology technique out there for you.
Jackson ImmunoResearch offers an extensive selection of high-quality reagents to support your research, and whichever antibody-based method like IHC, CycIF, and t-CyCIF you choose, the same fundamental principles apply. Read more to find out what considerations are important when developing your spatial biology experiment.
References
- Lin, J. R., Fallahi-Sichani, M., & Sorger, P. K. (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nature communications, 6, 8390. https://doi.org/10.1038/ncomms9390 https://pubmed.ncbi.nlm.nih.gov/26399630/
- Lin, J. R., Izar, B., Wang, S., Yapp, C., Mei, S., Shah, P. M., Santagata, S., & Sorger, P. K. (2018). Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife, 7, e31657. https://doi.org/10.7554/eLife.31657 https://pubmed.ncbi.nlm.nih.gov/29993362/
- Giesen, C., Wang, H. A., Schapiro, D., Zivanovic, N., Jacobs, A., Hattendorf, B., Schüffler, P. J., Grolimund, D., Buhmann, J. M., Brandt, S., Varga, Z., Wild, P. J., Günther, D., & Bodenmiller, B. (2014). Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature methods, 11(4), 417–422.https://pubmed.ncbi.nlm.nih.gov/24584193/
- Goltsev, Y., Samusik, N., Kennedy-Darling, J., Bhate, S., Hale, M., Vazquez, G., Black, S., & Nolan, G. P. (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell, 174(4), 968–981.e15. https://doi.org/10.1016/j.cell.2018.07.010 https://pubmed.ncbi.nlm.nih.gov/30078711/
- Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M., & Cai, L. (2014). Single-cell in situ RNA profiling by sequential hybridization. Nature methods, 11(4), 360–361. https://doi.org/10.1038/nmeth.2892 https://pubmed.ncbi.nlm.nih.gov/24681720/
- Eng, C. L., Lawson, M., Zhu, Q., Dries, R., Koulena, N., Takei, Y., Yun, J., Cronin, C., Karp, C., Yuan, G. C., & Cai, L. (2019). Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature, 568(7751), 235–239. https://doi.org/10.1038/s41586-019-1049-y https://pubmed.ncbi.nlm.nih.gov/30911168/
- Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S., & Zhuang, X. (2015). RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science (New York, N.Y.), 348(6233), aaa6090. https://doi.org/10.1126/science.aaa6090 https://pubmed.ncbi.nlm.nih.gov/25858977/
- Wang, F., Flanagan, J., Su, N., Wang, L. C., Bui, S., Nielson, A., Wu, X., Vo, H. T., Ma, X. J., & Luo, Y. (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics : JMD, 14(1), 22–29. https://doi.org/10.1016/j.jmoldx.2011.08.002 https://pubmed.ncbi.nlm.nih.gov/22166544/
- Gerlinger, M., Rowan, A. J., Horswell, S., Math, M., Larkin, J., Endesfelder, D., Gronroos, E., Martinez, P., Matthews, N., Stewart, A., Tarpey, P., Varela, I., Phillimore, B., Begum, S., McDonald, N. Q., Butler, A., Jones, D., Raine, K., Latimer, C., Santos, C. R., … Swanton, C. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine, 366(10), 883–892. https://doi.org/10.1056/NEJMoa1113205 https://pubmed.ncbi.nlm.nih.gov/22397650/
- Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B. R., Sabo, P. J., Dorschner, M. O., Sandstrom, R., Bernstein, B., Bender, M. A., Groudine, M., Gnirke, A., Stamatoyannopoulos, J., Mirny, L. A., Lander, E. S., & Dekker, J. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, N.Y.), 326(5950), 289–293. https://doi.org/10.1126/science.1181369 https://pubmed.ncbi.nlm.nih.gov/19815776/
- Steensel, B., Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat Biotechnol18, 424–428 (2000). https://doi.org/10.1038/74487 https://www.nature.com/articles/nbt0400_424
- Ahanger, S. H., Delgado, R. N., Gil, E., Cole, M. A., Zhao, J., Hong, S. J., Kriegstein, A. R., Nowakowski, T. J., Pollen, A. A., & Lim, D. A. (2021). Distinct nuclear compartment-associated genome architecture in the developing mammalian brain. Nature neuroscience, 24(9), 1235–1242. https://doi.org/10.1038/s41593-021-00879-5 https://pubmed.ncbi.nlm.nih.gov/34239128/
- Schulz, D., Zanotelli, V. R. T., Fischer, J. R., Schapiro, D., Engler, S., Lun, X. K., Jackson, H. W., & Bodenmiller, B. (2018). Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell systems, 6(1), 25–36.e5. https://doi.org/10.1016/j.cels.2017.12.001 https://pubmed.ncbi.nlm.nih.gov/29289569/
- Liu, M., Lu, Y., Yang, B., Chen, Y., Radda, J. S. D., Hu, M., Katz, S. G., & Wang, S. (2020). Multiplexed imaging of nucleome architectures in single cells of mammalian tissue. Nature communications, 11(1), 2907. https://doi.org/10.1038/s41467-020-16732-5 https://pubmed.ncbi.nlm.nih.gov/32518300/

| Learn more: | Do more: |
|---|---|
| Indirect and direct Western blotting | Exhibition Schedule |
| Chemiluminescence western blotting | Western blotting guide |
| An Introduction to Expansion Microscopy | ELISA guide |