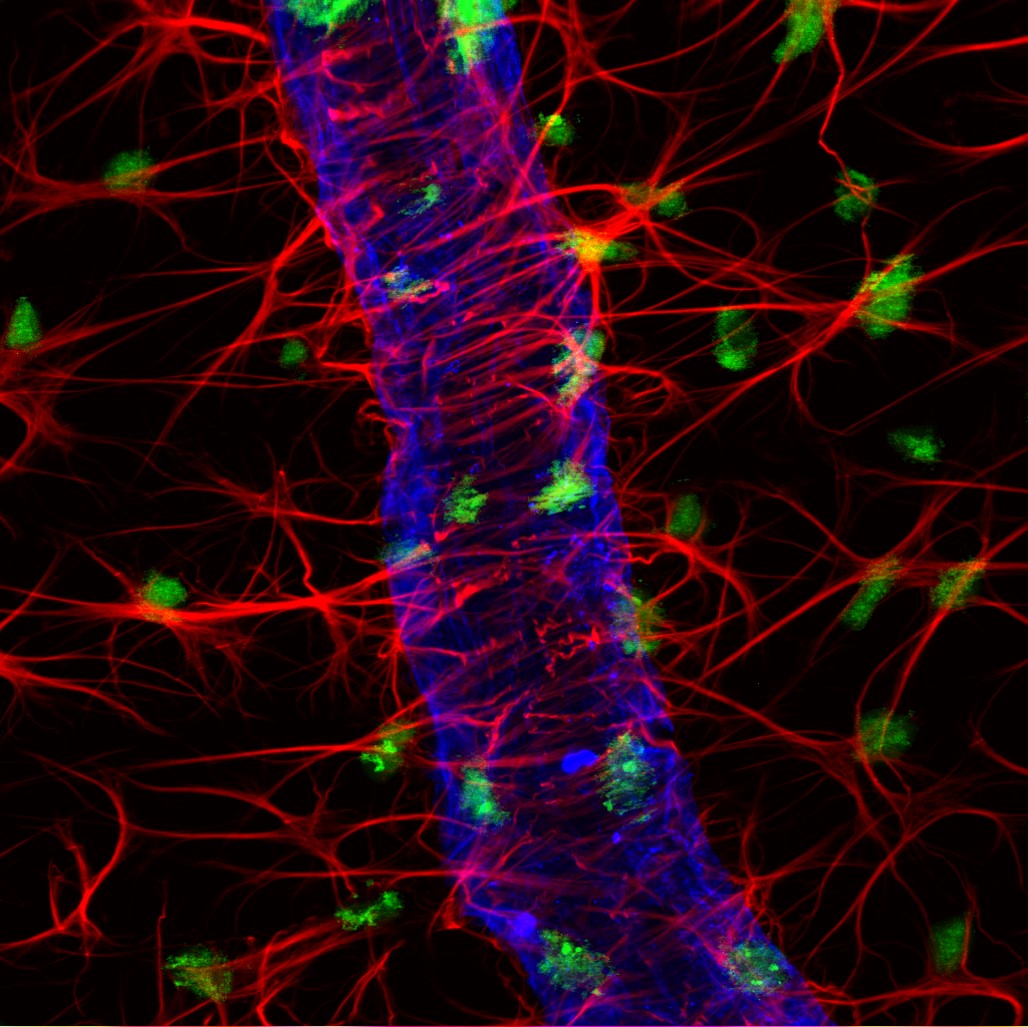
Expansion microscopy is a relatively new imaging technique that was developed to improve the resolution of conventional light microscopy without the need for dedicated instrumentation. It is seeing increased use for scientific research, as evidenced by a growing number of publications.
What is expansion microscopy?
Conventional microscopy has a resolution limit of approximately 200 nm, which is imposed by the diffraction of light as it passes through the instrument’s glass objective. This so-called ‘diffraction barrier’ prevents conventional microscopes from distinguishing between two objects that are separated by a distance that is less than half the wavelength of the imaging light source. Although this limitation can be overcome using more powerful microscopes (super-resolution microscopy), sophisticated algorithms, and smaller (often photoswitchable) fluorophores, the associated cost of advanced imaging techniques and the level of expertise required makes them inaccessible to many laboratories.
Expansion microscopy provides an alternative solution to the problem by increasing the resolution achievable with a conventional microscope. First described in 2015 by researchers at the Massachusetts Institute of Technology (MIT), expansion microscopy uses polyelectrolyte hydrogels to physically expand fixed cells and tissues. This is accomplished by exploiting the ability of polyelectrolyte hydrogels to gain or lose protons in response to changing pH (e.g., due to submersion in water), which causes them to swell. With expansion microscopy, volume increases of up to 100-fold, equating to around a 4.5-fold increase in the linear dimension, are commonly reported.
What are the main protocol steps for expansion microscopy?
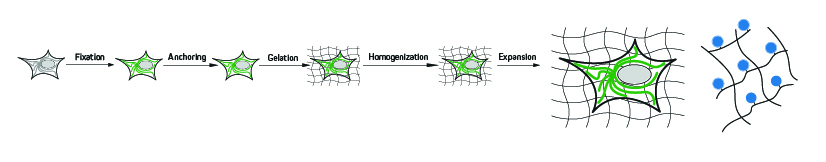
A typical expansion microscopy protocol follows several key steps, each of which must be carefully optimized to ensure the highest possible level of isotropy (sameness in all directions) and avoid introducing experimental artifacts. First, the cells or tissues are fixed before being treated with compounds that are capable of anchoring endogenous proteins to the hydrogel via covalent attachment. Next, a monomer solution of the hydrogel is added, which penetrates the sample and polymerizes to form a densely cross-linked structure – a process known as gelation. Following this, the sample is mechanically or enzymatically homogenized (depending on the sample material and the targets being visualized) to destabilize the proteins involved in maintaining cellular structure. It is then submersed in water, where it undergoes expansion.

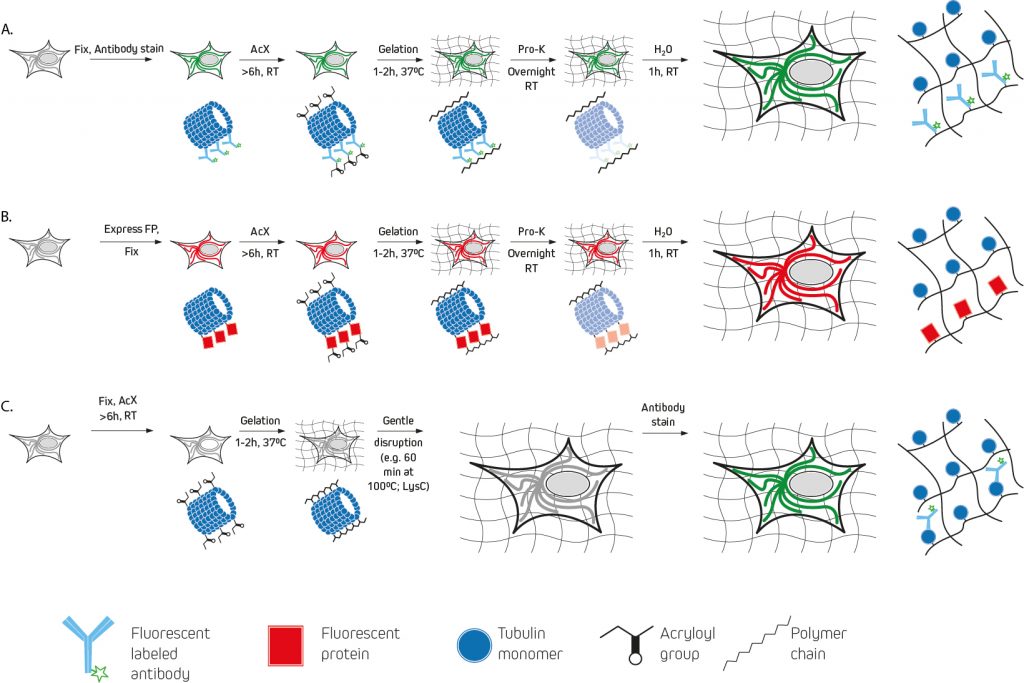
When are the labeled antibodies added?
The earliest expansion microscopy experiments used a custom label consisting of a methacryloyl group capable of participating in polymerization, a chemical fluorophore, and an oligonucleotide capable of hybridizing to a complementary sequence attached to an affinity tag (e.g., a secondary antibody) for pre-expansion labeling (figure 2:A). A drawback to this approach is that the DNA-labeled antibodies must be custom-made via a laborious and costly multi-step protocol. For this reason, other pre-expansion labeling methods have since been developed.
One of the best-known examples, protein retention expansion microscopy (proExM), uses the succinimidyl ester of 6-((acryloyl)amino)hexanoic acid (AcX), the methacrylic acid N-hydroxysuccinimidyl ester (MA-NHS), or glutaraldehyde for anchoring standard fluorophore-labeled antibodies (either labeled primary antibodies, or a combination of unlabeled primary antibodies and labeled secondary antibodies) to the polyelectrolyte hydrogel during polymerization. AcX and MA-NHS protocols have been developed for a wide range of fluorophore-labeled antibodies, and the MA-NHS method has also been adapted for use with biotinylated antibody reagents that can subsequently be detected using fluorophore-labeled streptavidin.
As an alternative to these methods, post-expansion labeling (figure 2:C) involves immunostaining samples only once the expansion process is complete.

Considerations for label selection
For post-expansion labeling protocols, almost any fluorophore-labeled antibody should be compatible, provided it is validated for immunocytochemistry (ICC) or immunohistochemistry (IHC). Likewise, most fluorophore-labeled antibodies are suitable for pre-expansion labeling, although it is important to note that some cyanine family dyes (e.g., Alexa Fluor® 647 and Cyanine 5) should not be used since they are destroyed during polymerization. If a far-red dye is needed, Alexa Fluor should be considered as an alternative.
Where expansion microscopy will be used for visualizing a recombinantly expressed fluorescent protein, most of these can survive the polymerization and expansion process. However, they may appear less bright due to the increased sample volume and correspondingly lower protein density. Infrared fluorescent proteins based on bacteriophytochromes are an exception, and it is advised that researchers use β-barrel structured proteins (e.g., green fluorescent protein) whenever possible.
Supporting expansion microscopy-based research
Jackson ImmunoResearch specializes in producing secondary antibodies for life science applications, including expansion microscopy. These are available conjugated to fluorescent proteins including R-phycoerythrin (R-PE), allophycocyanin (APC), and the peridinin-chlorophyll-protein complex (PerCP), as well as to synthetic fluorescent dyes including Alexa Fluor®, Brilliant Violet™, DyLight™, rhodamine, and fluorescein. We also offer fluorescently-labeled streptavidin, which can be used in combination with biotinylated antibodies to provide signal amplification.
References:
- Chen, F., Tillberg, P. and Boyden, E., 2015. Expansion microscopy. Science, 347(6221), pp.543-548.
- Gao, R., Asano, S.M. & Boyden, E.S. Q&A: Expansion microscopy. BMC Biol 15, 50 (2017). https://doi.org/10.1186/s12915-017-0393-3
- Gaudreau-Lapierre A, Mulatz K, Béïque JC, Trinkle-Mulcahy L. Expansion microscopy-based imaging of nuclear structures in cultured cells. STAR Protoc. 2021 Jun 26;2(3):100630. doi: 10.1016/j.xpro.2021.100630. PMID: 34223201; PMCID: PMC8243706.
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat Biotechnol. 2016 Sep;34(9):987-92. doi: 10.1038/nbt.3625. Epub 2016 Jul 4. PMID: 27376584; PMCID: PMC5068827.
- Wassie, A.T., Zhao, Y. & Boyden, E.S. Expansion microscopy: principles and uses in biological research. Nat Methods 16, 33–41 (2019). https://doi.org/10.1038/s41592-018-0219-4
[/noindex]


